Pneumonia Classification with PyTorch
This project builds a classifier for pneumonia detection in PyTorch from X-Ray images.
The RSNA Pneumonia detection challenge data is from ‘kaggle.com/c/rsna-pneumonia-detection-challenge/’. It is nearly 27,000 X-Ray images.
Per usual operation of Google Colab, mount to your Drive and then change directory to where your project files are held.
Here are a list of libraries needed with associated functionality:
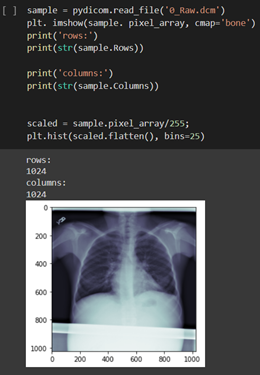
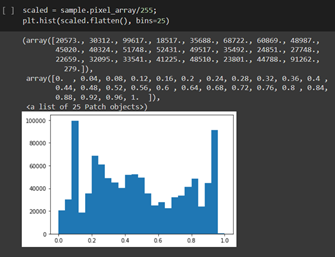
Viewing different intensities after simple scaling:
We invoke different methods for transforming/manipulating the images. Tilt, translation, different scales applied, and different sizing (cropping).
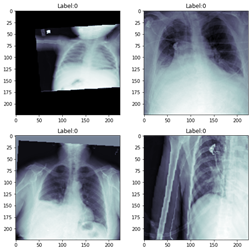
When we setup the loaders, we make very important decisions. Batch size, number of workers (consider your compute resources), number of images for training (24,000), and number of images for validation (2684). Later, we will vary the batch size parameter over a range of values during testing.
For training, we will use a number of additional libraries.
Without going into detail on the CNN formulation, I want to share a cool resource which puts a nice visual to the concept. Link: https://setosa.io/ev/image-kernels/
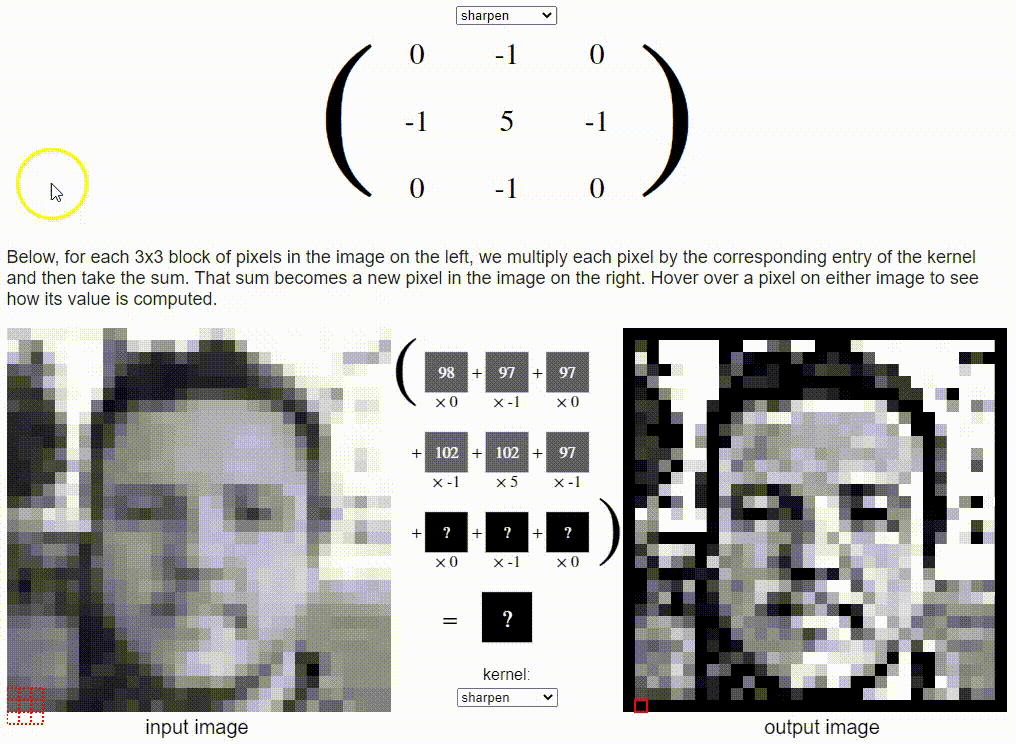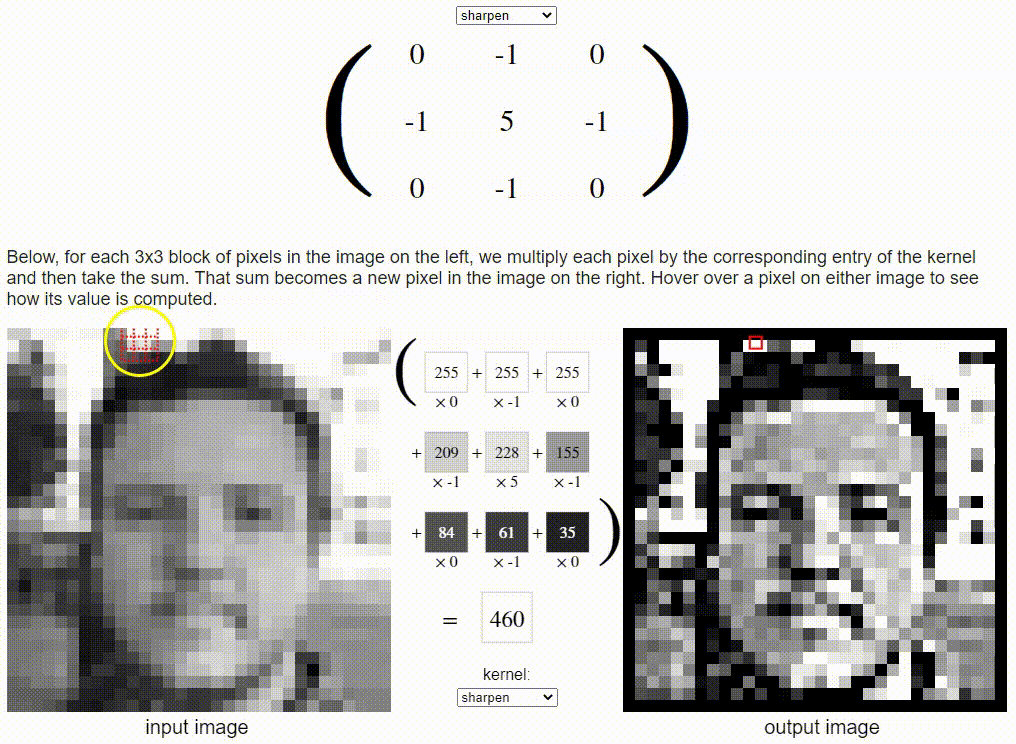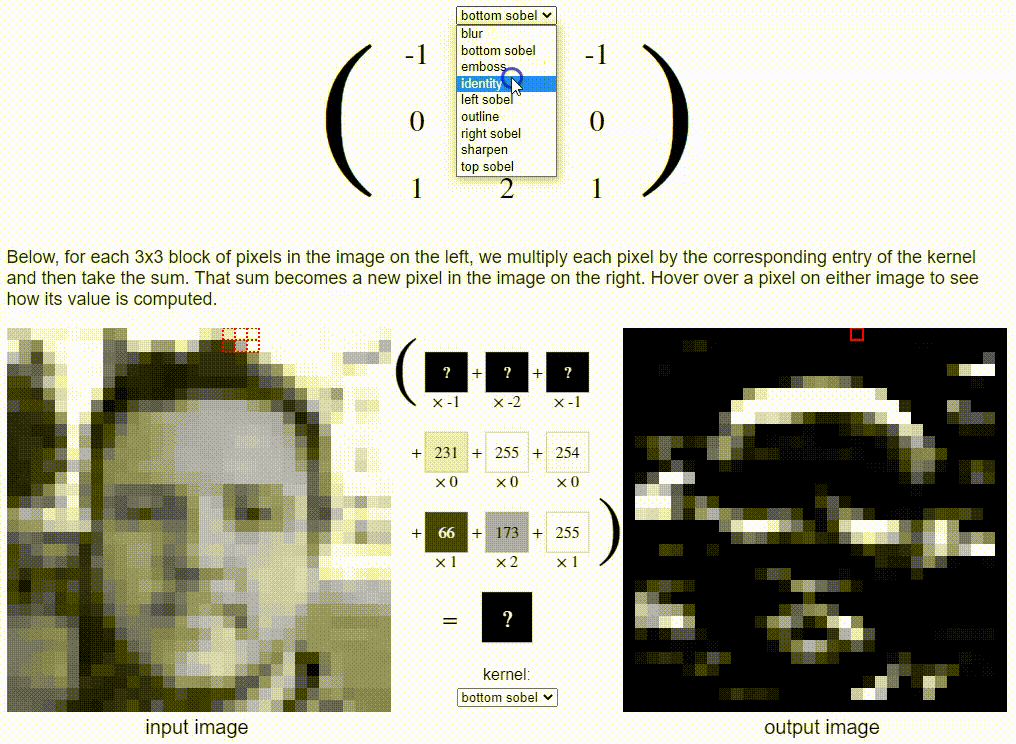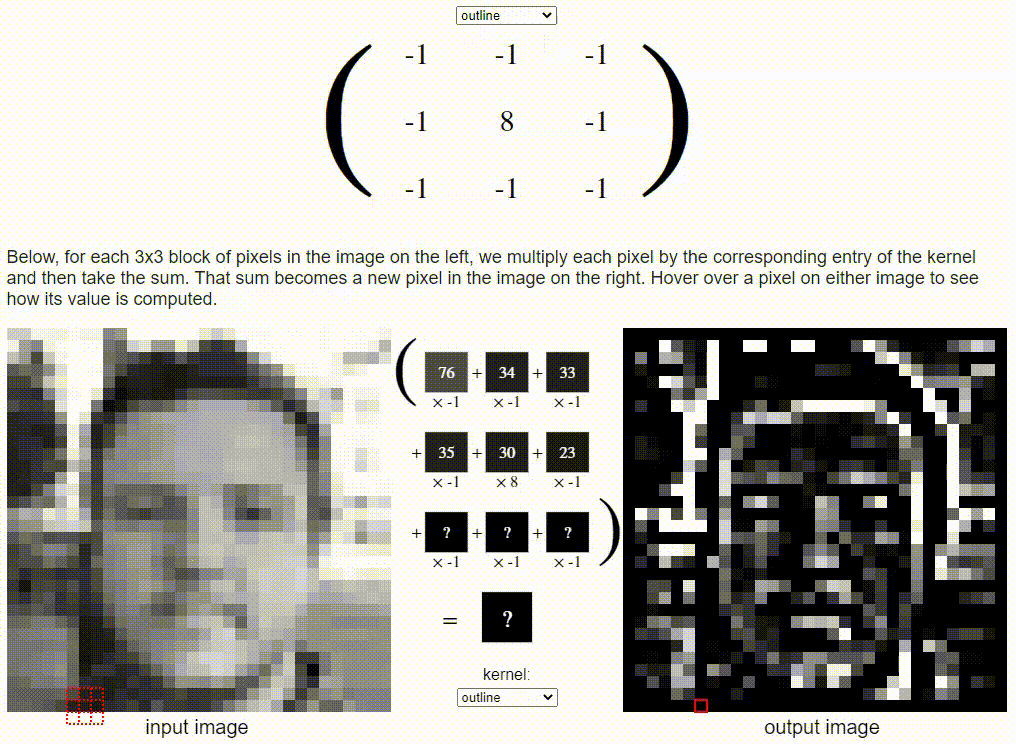
Two types of transfer learning for this project: finetuning and feature extraction. In finetuning, we start with a pretrained model and update all of the model’s parameters for our new task, in essence retraining the whole model. In feature extraction, we start with a pretrained model and only update the final layer weights from which we derive predictions. It is called feature extraction because we use the pretrained CNN as a fixed feature-extractor, and only change the output layer.
In general both transfer learning methods follow the same few steps:
- Initialize the pretrained model
- Reshape the final layer(s) to have the same number of outputs as the number of classes in the new dataset
- Define for the optimization algorithm which parameters we want to update during training
- Run the training step
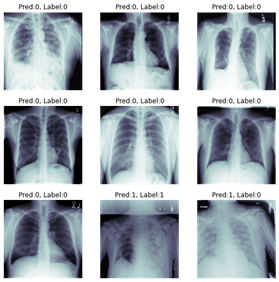
Showing a sample set of predictions once the model has been trained:
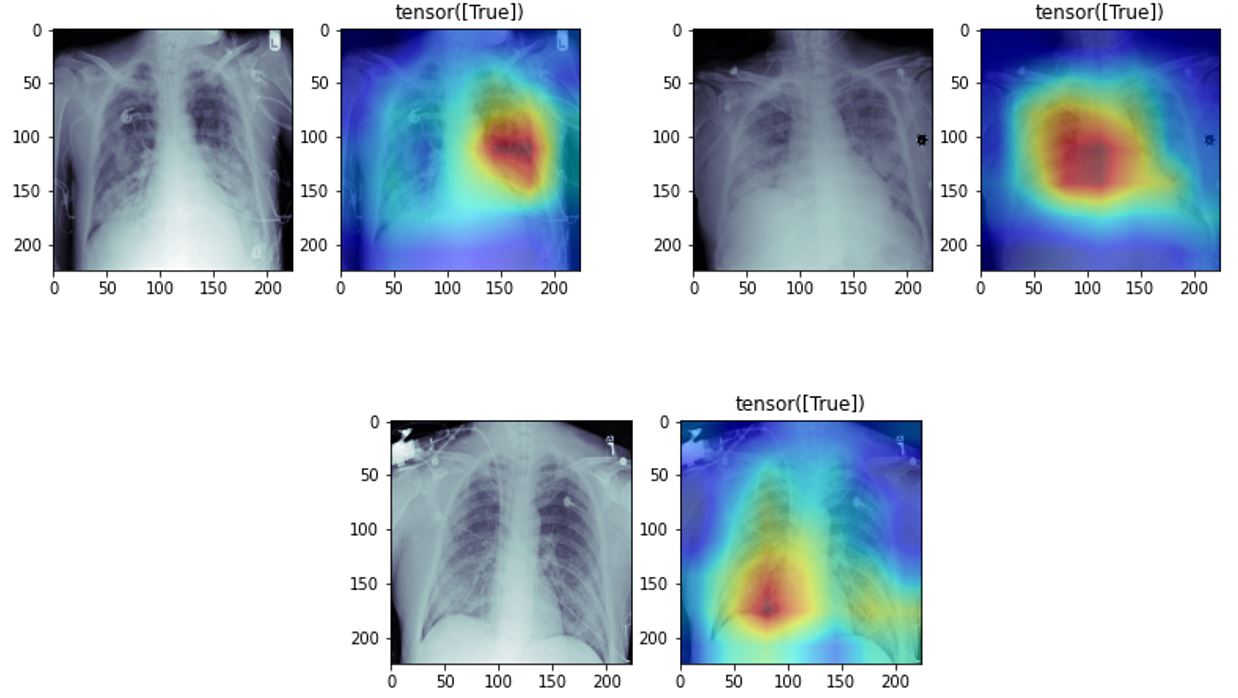
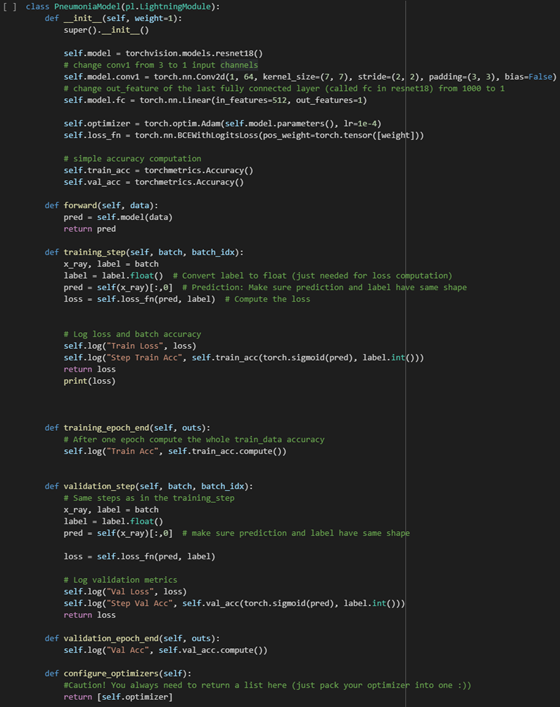
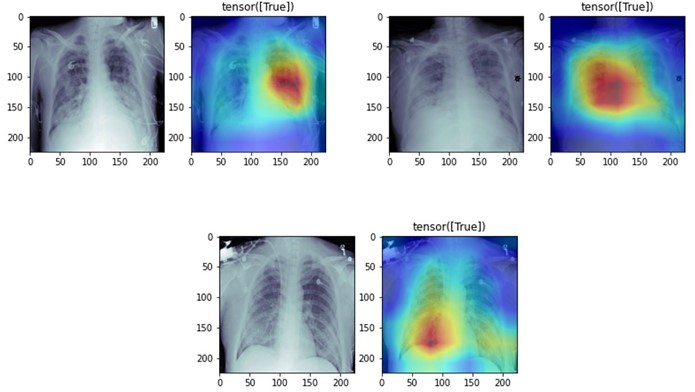
We can use a class activation map to discriminate which regions in the image used by the CNN contribute to class designation assigned.
Hyperparameter tuning is often described as an art more than a science, inefficient, and highly problem specific. I stumbled across a cool article that prescribed a way to help address this by focusing on batch size and learning rate first. It seeks to find a number of nearly flat directions in hyperparameter space. This greatly simplifies the search problem. Optimizing along these directions helps a little, but the penalty for doing so badly, is small.
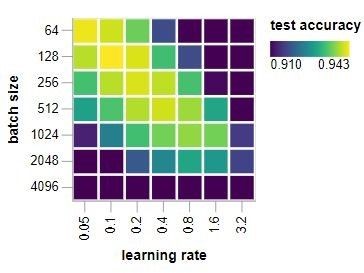
In the pneumonia classification study, this process boosted the validation accuracy to 0.8405 (+3%) and the validation precision .7025 (+6%). Several batch sizes were tested (16, 32, 64, 128, 256, 512, 1024) as well as different learning rates (0.0001, 0.0002) for a fixed number of epochs (100). In the end, I guess it’s no surprise that Yan LuCan’s words held true.

